Restriction digestion
Master mix preparation:
Prepare a master mix of the following per sample, plus 5 to 10% extra to allow for pipetting loss.
5X R/L buffer (see page 4 for recipe) 6.0 μl
EcoR I (12 U @20 U/μl) 0.6 μl
Mse I (8 U @ 4 U/μl) 2.0 μl
Water to 10 μl
Reaction time:
Incubate for at least 1 hr. at 37°C, but not longer than 3 hrs, before proceeding with the ligation.
Adapter ligation
Master mix preparation
If 10 μl of the restriction reaction was removed for gel analysis, prepare a ligation master mix of the following, per sample, plus enough for 5-10 extra samples:
EcoRI adapter (@ 5 pMol/μl) 0.5 μl
MseI adapter (@ 50 pMol/μl) 0.5 μl
ATP (10 mM, pH 8.0) 0.5 μl
5X R/L buffer 1.0 μl
sdH2O 2.0 μl
T4 DNA ligase (0.5 Weiss U @ 1 U/μl) 0.5 μl
TOTAL 5.0 μl
Reaction time:
Incubate for at least 3 hrs. (preferably overnight) @ 37° C. After incubation, dilute each R/L mix 1:10 with sdH2O.
Preamplification
Master mix preparation
Prepare a master mix with the following amounts per sample, plus 5-10 samples extra:
10X PCR buffer 3.0 μl
dNTP mixture (2.5 mM ea.) 2.4 μl
E primer (@ 50 ng/μl ≅ 8.3μM) 1.0 μl
M primer (@ 50 ng/μl ≅ 8.3μM) 1.0 μl
Taq polymerase (@ 5 U/μl) 0.4 μl
sdH2O 19.2 μl
μl per sample 27.0 μl
Important: If pre-amplifications are to be done in 96-well PVC plates instead of polycarbonate or polypropylene plates or tubes, it may be necessary to add 2.0 μl of 10 μg/μl BSA per sample to the Taq-buffer mix, and decrease the amount of H2O to 5.76 μl per sample. DO NOT use a hot start PCR protocol, or a hot start polymerase (e.g. AmpliTaq Gold), for the pre-amplification! The adapters are non-phosphorylated, so only the top strand is ligated. The bottom strand of the adapter will separate from the rest of the template first and must be re-synthesized during the initial heating stage. This requires polymerase activity during the initial heating.
PCR program
Place reactions in the thermocycler, and run the following PCR amplification profile:
28 cycles: 15 sec @ 94°C denaturation
30 sec @ 60°C annealing
60 sec + 1sec/cycle @ 72°C extension
1 cycle: 2 min @ 72°C final extension
hold: 4°C
Final Amplification
Master mix preparation (single dye reactions)
For each E/M primer combination to be used, prepare the following master mix, per sample, in an Eppendorf tube, plus enough for 5-10 samples extra:
10X PCR buffer 2.0 μl
dNTP mixture (2.5 mM ea) 1.6 μl
IRD-labeled E-primer (@ 6 ng/μl ≅ 1μM) 0.83 μl
M-primer (@ 50 ng/μl ≅ 8.3μM) 0.6 μl
Taq polymerase (@ 5U/μl) 0.24 μl
dH2O 9.73 μl
Final volume 15.0 μl
PCR program
Amplify in a thermocycler equipped for microtiter plates, using the following PCR profile:
13 cycles: 10 sec @ 94°C denaturation
30 sec @ 65°C annealing, less 0.7° per cycle after the first cycle
60 sec @ 72°C extension
25 cycles: 10 sec @ 94°C denaturation
30 sec @ 56°C annealing
60 sec @ 72°C extension, plus 1 sec. per cycle
1 cycle: 2 min @ 72°C final extension
hold: 4°C
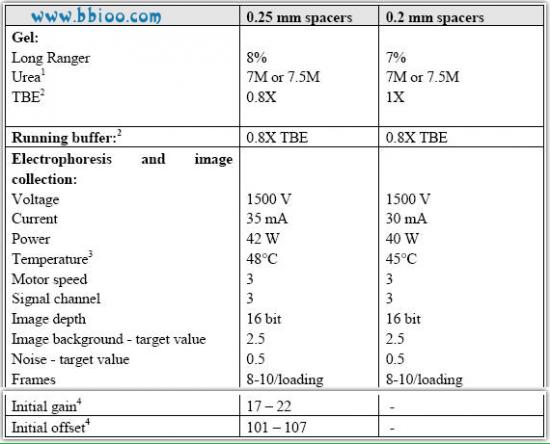
Gel and automated electrophoresis conditions
Gel and electrophoresis conditions for running AFLP reactions on LI-COR automated sequencers(models 4000 and 4200).
1 Use only ultra pure grade urea. Gel solutions with 7M urea tend to produce less precipitate at 4°C.
2 The concentration of TBE in the gel solution and running buffer was reduced from 1.0X to 0.8X to reduce the current (mA) and prevent overheating during the run. LI-COR automated sequencers do not have active cooling capability, and the heat production inside the gels sometimes cause temperatures to rise to between 53°C and 56°C by the end of the runs (this is a problem during the summer when temperatures can rise to approx. 26°C in our laboratory). Overheating causes the gels to break up at the end of the first runs and prevent us from loading second runs on the gels.
3 We start the running temperature at 48°C to prevent "overshooting" 50°C by the end of the run.
4 The initial gain and offset values given here are only appropriate for version 4000 of the Base ImagIR data collection software (used on single-dye, IRD800 sequencers) that requires manual setting of the gain and offset parameters to obtain the target background and noise values.
The newer version 4200 of the software is able to find the appropriate gain and offset values during the Auto gain procedure.
Buffers:
5X restriction-ligation (R/L) buffer
In addition to 1M Tris HAc pH 7.5 (see above), prepare the following solutions:
1M Mg acetate (MgAc): To 2.145 g (tetrahydrate - mol. wt. 214.5 gr/mol), add dH2O to 10 ml. Filter sterilize.
1M K acetate (KAc): To 0.981 g, add sdH2O to 10 ml. Store at -20°.
1M DTT: To 1.542 g, add sdH2O to 10 ml. (DTT is very loose and sticky;a large funnel may be useful for transferring to a jar or conical tube. Store at -20°.)
Prepare 5X R/L buffer as follows:
50 mM Tris HAc pH 7.5 0.5 ml of 1M
50 mM MgAc 0.5 ml of 1M
250 mM KAc 2.5 ml of 1M
25 mM DTT 250 μl of 1M
sdH2O 6.0 ml (to 9.75 ml total volume)
Store at -20°C in 975 μl aliquots. Upon use, add 25 μl of 10 mg/ml BSA (e.g. NE Biolabs) to a single aliquot, for 250 ng/μl final concentration. Store unused portion at 4°C.
Adapters:
EcoRI adapter:
(5 pMol/μl): 5'-CTC GTA GAC TGC GTA CC
CAT CTG ACG CAT GGT TAA-5'
In 0.5-ml microfuge tube, prepare the following:
top strand (1μg/μl) 8.5 μl (approx. 1500 pMol)
bottom strand (1μg/μl) 9.0 μl (approx. 1500 pMol)
sdH2O 282.5 μl
300.0 μl
In a thermocycler, heat to approx. 90° for 2-3 min, then cool gradually (at e.g. 3°C/min) to room temperature. Store at -20°.
MseI adapter:
(50 pMol/μl): 5'-GAC GAT GAG TCC TGA G
TA CTC AGG ACT CAT-5'
In 0.5-ml microfuge tube, prepare the following:
top strand (1μg/μl) 80 μl (approx. 15,000 pMol)
bottom strand (1μg/μl) 70 μl (approx. 15,000 pMol)
sdH2O 150 μl
300 μl
In thermocycler, heat to approx. 90° for 2-3 min, then cool gradually (at e.g. 3°C/min) to room temperature. Store at -20°.
Primers:
The core primer sequences (without selective extensions) are:
EcoRI primer (E-primer) 5'-GAC TGC GTA CCA ATT C-3'
MseI primer (M-primer) 5'-GAT GAG TCC TGA GTA A-3'
* An additional “A” at the 3’ end of the E-primer gives an optimum number of bands for Salmonella fingerprinting purpose. I have not tried other bases but others may give a better result as well.
Gel solutions and buffers
(a) (a) Long Ranger gel solution:
It is most convenient to prepare 500 ml a stock gel solution ahead of time. The following is for a 8% Long Ranger gel solution with 0.8X TBE; adjust Long Ranger and TBE accordingly for other concentrations:
7.0 M urea 210.2 g
0.8 X TBE 40 ml of 10X (or 50ml of 8X)
8% Long Ranger acrylamide 80 ml of 50% (from FMC)
dH2O to 500 ml final volume
Weigh the urea (Ultra pure urea from AMRESCO) into a 1-L beaker. Wearing appropriate protective clothing (DANGER: acrylamide is a potent neurotoxin until it is polymerized!!), add the TBE and Long Ranger to the urea in the beaker. Add dH2O to slightly less than 500 ml volume, and stir under low heat (setting 1-2 on heater-stirrer)
until dissolved. Pour carefully into a graduated cylinder and add dH2O to 500 ml. Filter through a 0.2μm filter using vacuum. Store at 4°C or room temperature in a foil-covered bottle. If stored at 4°, re-dissolve any precipitated urea before using. Notes: Do not let the gel solution get warmer than room temperature while dissolving the urea. If enough
time is available, do not use external heat, but let the solution stir for a couple of hours until completely dissolved.
10X TBE:
108 g Tris base
55 g boric acid
7.44 g EDTA (disodium salt) or 40 ml 0.5 M EDTA pH 8.0
Add 800 ml dH2O, and stir on stir plate until dissolved. Then add dH2O to 1L.
10% ammonium persulfate (APS):
To 1 g APS, add sdH2O to 10 ml. Store at -20°C in 200 μl aliquots. Discard used tubes.
Formamide loading dye/stop buffer (for automated sequencer analysis):
Before making loading dye, it is best to deionize the formamide. Place about 2.5 g.mixed-bed resin beads (e.g. BioRad AG501-X8) in a small beaker. Add 2 to 3 ml formamide, swirl and discard the formamide. (A P1000 pipettor works well to remove the formamide.) Then add 50 ml formamide, and stir ~20 min on a stir plate. Filter the formamide through Whatman filter paper set in a vacuum funnel.
Deionize formamide as described above, and prepare the buffer as follows:
95% formamide 47.5 ml formamide
20 mM EDTA 2 ml of 0.5 M EDTA
sdH2O 0.5 ml
bromophenol blue (e.g. USB) ~40 mg
Note: At least some lots of Sigma bromophenol blue appear to quench the IRD signal, so use with caution. IR2 stop buffer, with basic fuchsin dye, is not necessary for marker applications, as the dye front will migrate below the first fragments and will not interfere with their detection.
Low BFB loading dye:
15 % Ficoll - Weigh 7.5 gr Ficoll (type 400) into a 100-ml bottle. Add water to approx.45 ml. Heat briefly in a microwave at low heat setting. Close lid and shake vigorously.Repeat heating and shaking until completely dissolved (NB. remove lid when heating in microwave). Add dH2O to 50-ml mark.
0.05% bromophenol blue (BFB) - Add 25 mg BFB to dissolved Ficoll, close lid and shake well.
Notes: This loading dye can be stored at room temperature. The purpose of adding less BFB (0.05% in stead of 0.25% commonly used) is to eliminate the dark blue band that usually migrates at about 500 bp in 0.8% to 1.2% gels. This helps to better visualize restriction digest and preamplification smears that run from 0.2kb to 1.0 kb.