基因的重复和分化为生物的演化提供了原材料。重复基因的分化可以发生在调控区或者编码区。发生在编码区的、能够导致功能分化的突变大体上分为两类,即非同义替换和内含子-外显子结构变化,前者导致同源位点上的氨基酸替换,后者则引起氨基酸的插入和缺失。作为点突变的一种方式,非同义替换的机制、速度和后果都已比较清楚。但是,相对而言,人们对内含子-外显子结构分化的研究尚不深入。
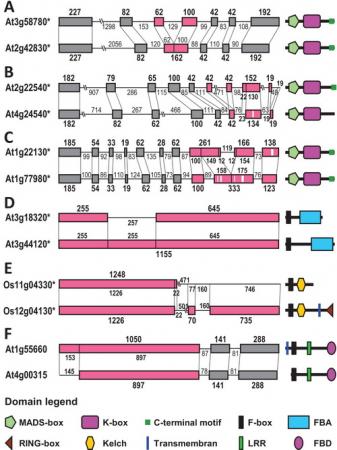
图片说明:重复基因结构分化的多种类型及结构域变化。图中左边为内含子-外显子结构比较示意图,右边为相应的蛋白质结构域组成式样。灰色和红色方框分别表示未发生分化和发生了分化的外显子,红色方框中用白色竖线标示出了插入/缺失事件发生的位置。
继2007和2009年发现内含子-外显子结构分化对重复基因分化的贡献之后,系统与进化国家重点实验室孔宏智研究组对这一问题进行了进一步的研究。通过深入分析612对重复基因(即旁系同源基因)和300对非重复基因(即直系同源基因),该研究组不但再次确认了内含子-外显子结构分化在重复基因分化中的普遍性和重要性,而且发现了导致结构分化的三类主要机制(即插入/缺失、外显子化/假外显子化和外显子/内含子获得/丢失)发生的频率及其对结构和功能分化的作用各不相同。
有意思的是,与分化时间大致相当的非重复基因相比,重复基因经历和积累了明显更多的结构分化,说明结构分化对重复基因分化的贡献更大,是导致新功能和新基因快速产生的主要原因。
该研究对于理解基因进化的基本式样以及新功能、新基因和新性状产生的分子机制等基本生物学问题具有重要意义。
上述研究结果于1月9日在《美国国家科学院院刊》上在线发表。该研究得到国家科技部和国家自然科学基金委的资助。

Divergence of duplicate genes in exon–intron structure
Guixia Xu, Chunce Guo, Hongyan Shan, and Hongzhi Kong
Gene duplication plays key roles in organismal evolution. Duplicate genes, if they survive, tend to diverge in regulatory and coding regions. Divergences in coding regions, especially those that can change the function of the gene, can be caused by amino acid-altering substitutions and/or alterations in exon–intron structure. Much has been learned about the mode, tempo, and consequences of nucleotide substitutions, yet relatively little is known about structural divergences. In this study, by analyzing 612 pairs of sibling paralogs from seven representative gene families and 300 pairs of one-to-one orthologs from different species, we investigated the occurrence and relative importance of structural divergences during the evolution of duplicate and nonduplicate genes. We found that structural divergences have been very prevalent in duplicate genes and, in many cases, have led to the generation of functionally distinct paralogs. Comparisons of the genomic sequences of these genes further indicated that the differences in exon–intron structure were actually accomplished by three main types of mechanisms (exon/intron gain/loss, exonization/pseudoexonization, and insertion/deletion), each of which contributed differently to structural divergence. Like nucleotide substitutions, insertion/deletion and exonization/pseudoexonization occurred more or less randomly, with the number of observable mutational events per gene pair being largely proportional to evolutionary time. Notably, however, compared with paralogs with similar evolutionary times, orthologs have accumulated significantly fewer structural changes, whereas the amounts of amino acid replacements accumulated did not show clear differences. This finding suggests that structural divergences have played a more important role during the evolution of duplicate than nonduplicate genes.
文献链接:https://www.pnas.org/content/early/2012/01/04/1109047109.abstract








